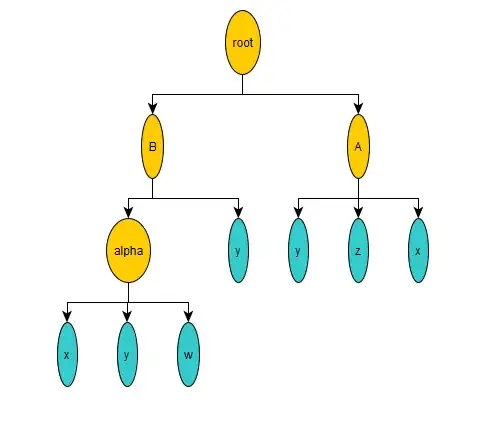
In my work I frequently come across systems of interdependent equations. I have contrived a toy example as follows. The terminal values w, x, y and z are given:
e(y) = A+B
A(y) = x*log(y)+y^z
B(y) = alpha*y
alpha(y) = x*y+w
We could then consider the function e(y) as the root of an arithmetic tree with the following heirarchy:
Previously, in python I would have done something like this to evaluate the result:
import numpy as np
def root(B, A):
return B+A
def A(x,y,z):
return x*np.log(y)+y**z
def B(alpha, y):
return alpha*y
def alpha(x,y,w):
return x*y+w
if __name__=='__main__':
x,y,z,w = 1,2,3,4
result = root(B(alpha(x,y,w),y), A(x,y,z))
This will give me the right result, but I have come to really despise this way of doing things. It requires me to keep track of which arguments each function needs and how the tree itself is built up. Also, suppose I wanted to modify the tree itself by adding branches and leaves. For example, say I wanted to redefine alpha as v+x+y with the new variable v. I'd have to make a new function and a new call, which is not very efficient as I sometimes need to make pervasive and numerous changes.
I tried different approaches to solve this problem as outlined by this question and this question.
I came across a couple of ideas which looked promising, namely function objects and the Interpreter Pattern. However I was disappointed by the Interpreter Pattern. Suppose I didn't create a parser, and went straight for the underlying composite architecture, wouldn't I still have to do something like this?
root = root_obj(B_obj(alpha_obj(x_obj,y_obj,w_obj),y_obj), A(x_obj,y_obj,z_obj))
root.interpret()
The above would require a lot of added complexity for no added value. My question is as follows: What is a simple and useful object oriented paradigm in which I could define, modify and evaluate a mathematical heirarchy in a dynamic manner?
EDIT
Here's an example of what I would like to achieve:
tree = FunctionTree()
tree.add_nodes(root, A, B, alpha, w, x, y, z)
tree.add_edge(root, [A, B])
tree.add_edge(root, A)
tree.add_edge(A, [x,y,z])
tree.add_edge(B, [alpha, y])
tree.add_edge(alpha, [x, y, w])
tree.evaluate()
Yes, this is less "compact" but it is much more flexible. Imaging having methods for deleting and adding new edges. replacing definitions at nodes and reevaluating the result. I am looking for something like this.