Currently I have a large ontology like this:
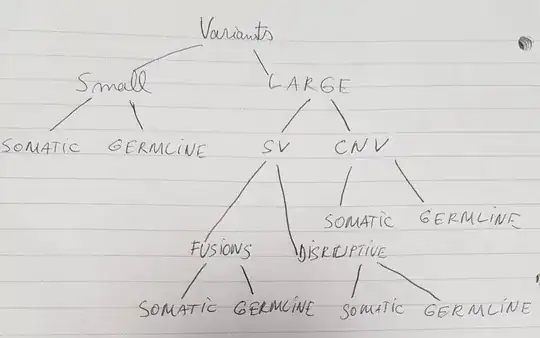
There are different categories of variants and each can be somatic or germline. There are more common behaviours between same variants, like somatic CNVs and germline CNVs have more common behaviours due to the fact that they are both CNVs. But there also are a few behaviours common between small somatic and somatic fusions due to them being somatic (this design doesn't account for that kind on reuse).
So I'm thinking of changing it to something else, a strategy pattern with a factory, like this:

But this runs into the issue that I have too many behaviours and they are quite specific to a class of variant. Example of behaviours:
- calculate_position()
- calculate_gene_location()
- calculate_log2foldchange()
All these do something different depending on variant type. And they are quite short functions. So I'll end up creating a lot of small behaviour classes and end up with a more extensive ontology.
Not all variants have the same behaviours: some have a protein_prediction() or split_reads(). At the end of the day I want to be able to get 8 classes with the behaviours that I need.
Any ideas of a better way to organise my code?